Li, Yang, Dr.
Kontakt
Professur für Tierernährung
Eschikon 27
8315
Lindau
Switzerland
Research Area
Yang Li is a dedicated biochemist specializing in the fascinating realm of methanogen and rumen microbiome interaction. With a strong background in microbiology and bioinformatics, Yang's passion lies in finding innovative ways to mitigate greenhouse gas emissions and other pollutants originating from the agricultural sector. His ultimate goal is to create a more sustainable world for future generations to thrive in. Currently, Yang serves as a valuable member of the Animal Nutrition team within the Department of Environmental Systems Science at the renowned Agrovet-Strickhof research station. In this role, he spearheads vital in vitro experiments that contribute to our understanding of rumen systems and pave the way for impactful change.
Curriculum Vitae
After earning his doctorate from Massey University in collaboration with AgResearch Ltd, he embarked on an interesting postdoc in the laboratory of Professor Keith Ireton, unraveling the mysteries of of Listeria monocytogenes pathogenesis. His research expertise led him back to AgResearch, where he was hired as scientist to develop a high-throughput anti-methanogen assay, a key milestone in the methane mitigation METHLAB project. He later ventured to Switzerland, the home country of his wife, where he embarked on another captivating postdoc endeavor under the guidance of Emeritus Professor Michael Kreuzer in the team of Animal Nutrition. Currently, he is immersed in multiple cutting-edge in vitro projects under the guidance of Assistant Professor Mutian Niu, pushing the boundaries of scientific discovery.
Publications:
Li, Y., Crouzet, L., Kelly, W.J., Reid, P., Leahy, S.C. and Attwood, G.T. (2023) “Methanobrevibacter boviskoreani JH1T growth on alcohols allows development of a high throughput bioassay to detect methanogen inhibition.” Cur Res Microb Sci, p.100189, DOI: 10.1016/j.crmicr.2023.100189.
Zhang, X., Li, Y., Terranova, M., Ortmann, S., Kehraus, S., Gerspach, C., Kreuzer, M., Clauss, M. and Hummel, J. (2023). A pilot investigation on the effect of induced saliva flow on digestive parameters in sheep, and a comparison with cattle J Anim Physiol Anim Nutr., online, DOI: 10.1111/jpn.13815.
Zhang, X., Li, Y., Terranova, M., Ortmann, S., Kehraus, S., Gerspach, C., Kreuzer, M., Hummel, J. and Clauss, M. (2022) “Effect of induced saliva flow on fluid retention time, ruminal microbial yield and methane emission in cattle.” J Anim Physiol Anim Nutr., 107 (3):769-782, DOI: 10.1111/jpn.13773.
Zhang, X., Li, Y., Terranova, M., Ortmann, S., Kehraus, S., Gerspach, C., Kreuzer, M., Clauss, M. and Hummel, J. (2022) “Stimulating saliva flow in sheep: effects on variables of digestion, and a comparison with effects in cattle.” GfE conference 2022, Göttingen.
Li, Y., M. Kreuzer, Q. Clayssen, M.-O. Ebert, H.-J. Ruscheweyh, S. Sunagawa, C. Kunz, G. Attwood, S. L. Amelchanka · M. Terranova. (2021) “The rumen microbiome inhibits methane formation through dietary choline supplementation” Sci Rep, 11: 21761, DOI:10.1038/s41598-021-01031-w.
Dowd, G. C., R. Mortuza, M. Bhalla, V. N. Hoan, Y. Li, L. A. Rigano, and K. Ireton (2020). “Listeria monocytogenes exploits host exocytosis to promote cell-to-cell spread” PNAS, 117: 7, DOI: 10.1073/pnas.1916676117.
Li, Y., M. Kreuzer, C. Kunz and M. Terranova (2020) “Near complete reduction of rumen methane emission in vitro by supplementing high doses of choline” EAAP conference book of abstracts, p349.
Leahy, S. C., N. Doyle, P. Mbandlwa, G. T. Attwood, Y. Li, P. Ross and C. Stanton (2019). “Use of Lactic Acid Bacteria to Reduce Methane Production in Ruminants, a Critical Review” Front Microbiol., 10: 2207, DOI: 10.3389/fmicb.2019.02207.
Li, Y., W. Kelly, G. Attwood, P. Reid, and S. C. Leahy (2019). “Methanobrevibacter boviskoreanii JH1T growth on alcohols allows development of a high throughput bioassay” Congress on Gastrointestinal Function conference proceedings, 2019, p41. external pagehttps://www.congressgastrofunction.org/proceedings/2019_Congress_on_GI_Function.pdfcall_made
Kamke, J., P. Soni, Y. Li, S. Ganesh, W. J. Kelly, S. C. Leahy, W. Shi, J. Froula, E. Rubin and G. T. Attwood (2017). “Gene and transcript abundances of bacterial type III secretion systems from the rumen microbiome are correlated with methane yield in sheep.” BMC Res Notes, 10(1): article 367, DOI: 10.1186/s13104-017-2671-0.
Kamke, J., S. Kittelmann, P. Soni, Y. Li, M. Tavendale, S. Ganesh, P. H. Janssen, W. Shi, J. Froula, E. M. Rubin and G. T. Attwood (2016). “Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation.” Microbiome, 4(1): article 54, DOI: 10.1186/s40168-016-0201-2.
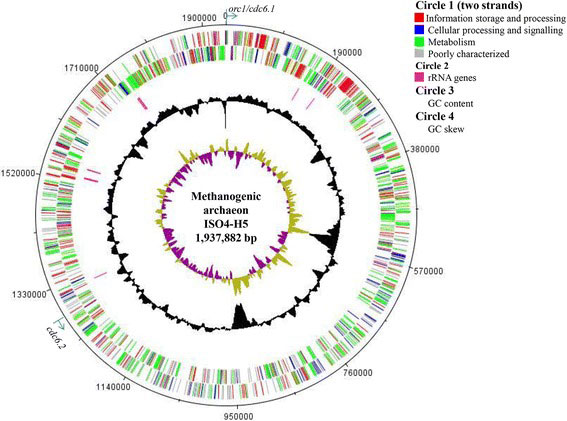
Li, Y., S. C. Leahy, J. Jeyanathan, G. Henderson, F. Cox, E. Altermann, W. J. Kelly, S. C. Lambie, P. H. Janssen and G. T. Attwood (2016). “The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales.” Stand Genomic Sci 11(1): article 59, DOI 10.1186/s40793-016-0183-5.
Kelly, W. J., D. Li, S. C. Lambie, J. Jeyanathan, F. Cox, Y. Li, G. T. Attwood, E. Altermann and S. C. Leahy (2016). “Complete genome sequence of methanogenic archaeon ISO4-G1, a member of the Methanomassiliicoccales, isolated from a sheep rumen.” Genome Announc 4(2): e00221-16.
Li, Y., R. Mortuza, D. L. Milligan, S. L. Tran, U. Strych, G. M. Cook and K. L. Krause (2014). “Investigation of the essentiality of glutamate racemase in Mycobacterium smegmatis.” J Bacteriol 196(24): 4239-4244.
Leahy, S. C., W. J. Kelly, D. Li, Y. Li, E. Altermann, S. C. Lambie, F. Cox and G. T. Attwood (2013). “The complete genome sequence of Methanobrevibacter sp. AbM4.” Stand Genomic Sci 8(2): 215-227.
